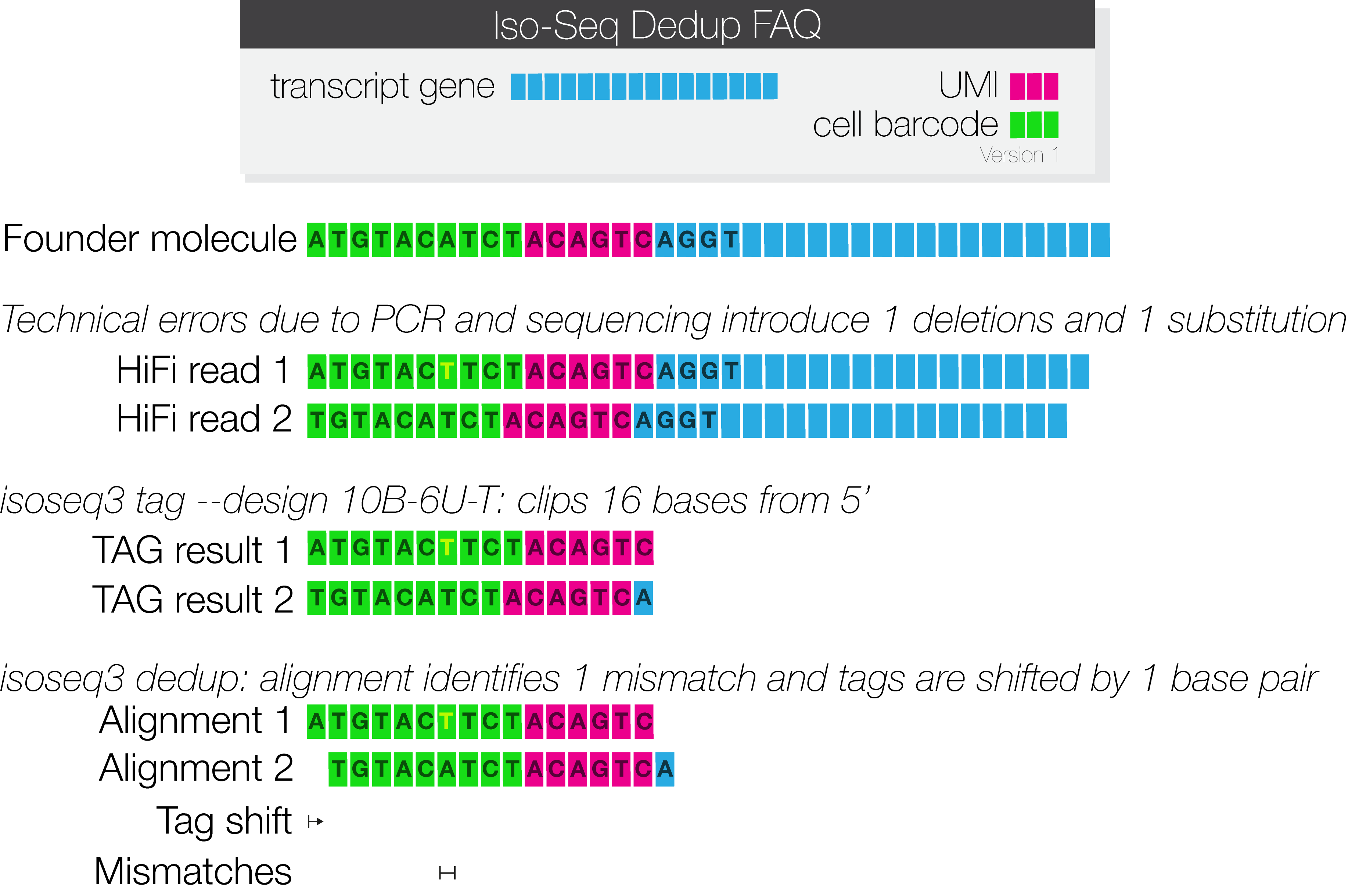
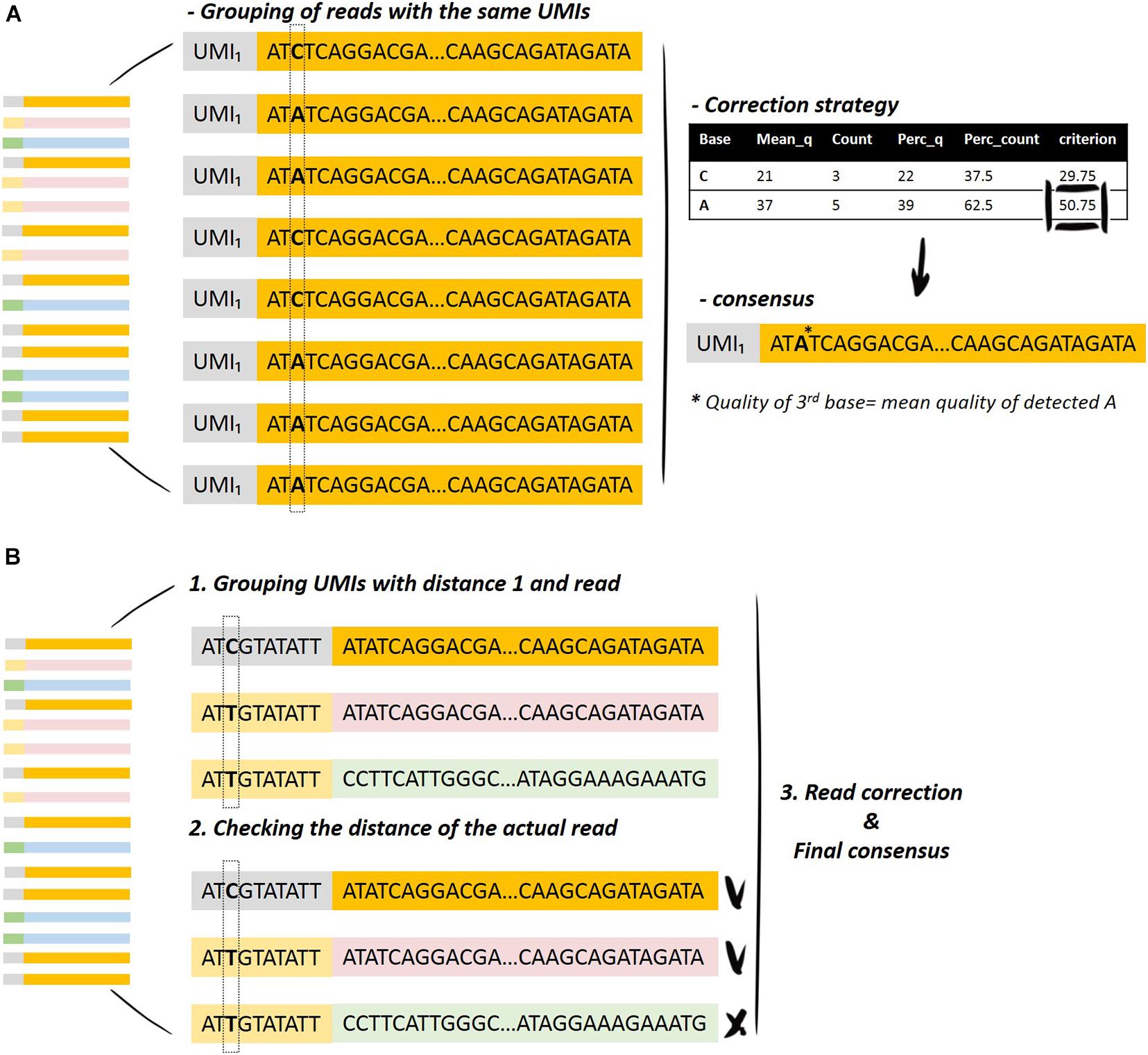
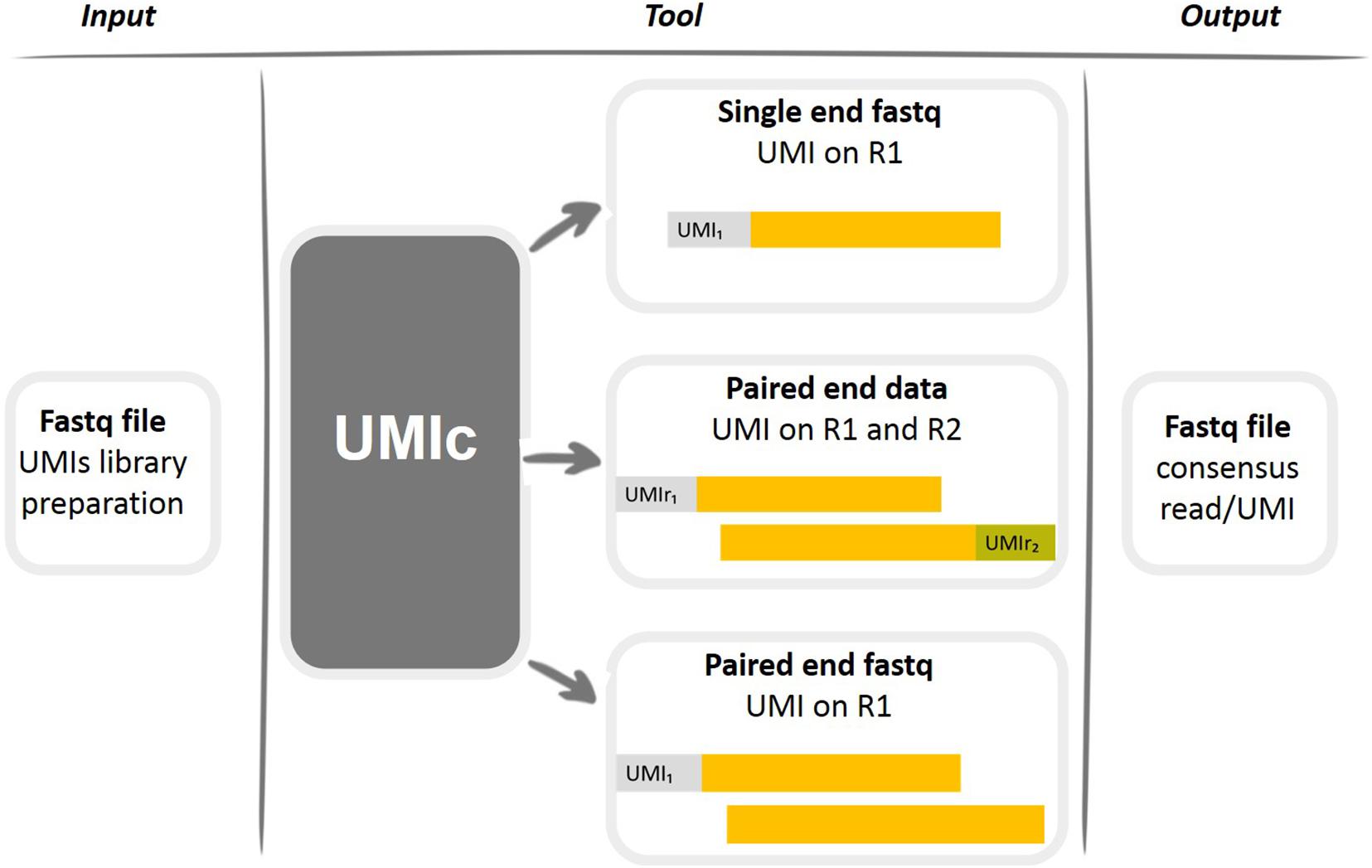
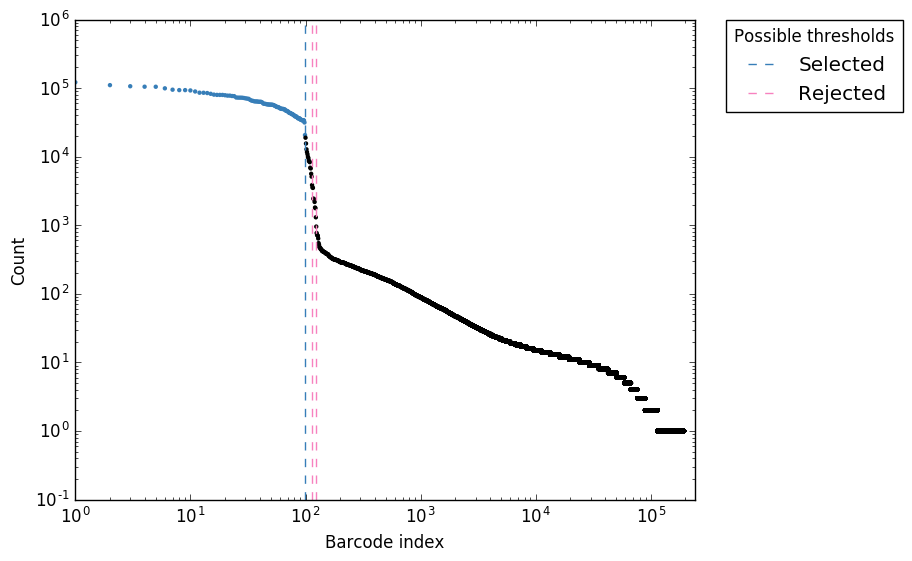
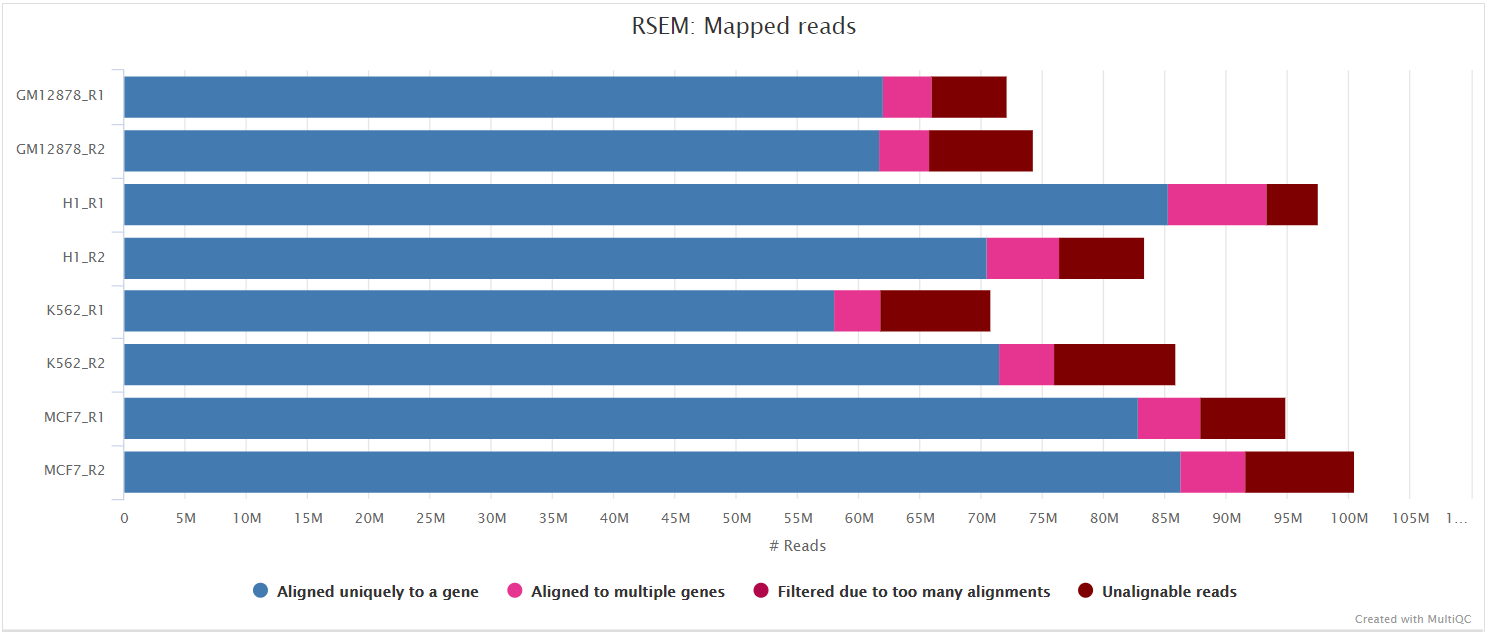
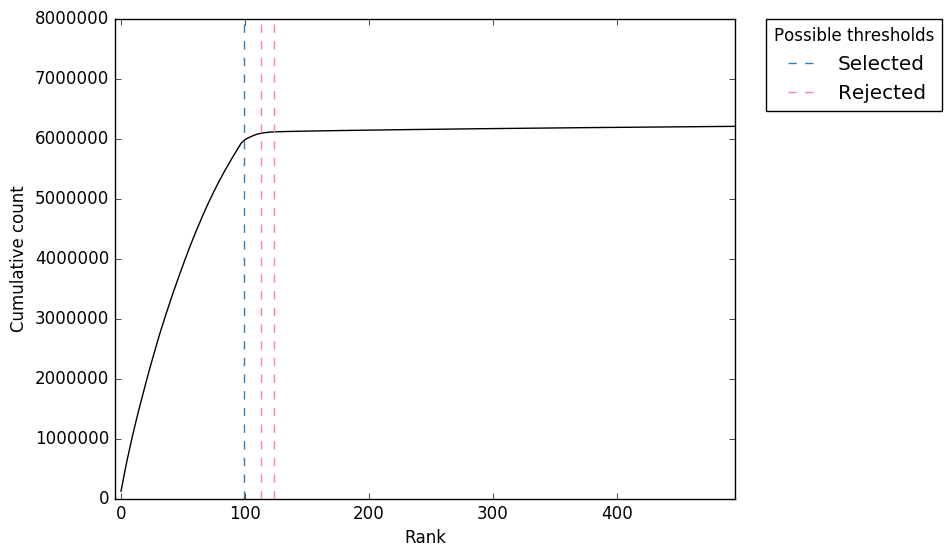
Analysis of UMIs. a Stacked bar plot showing the fractions of unique... | Download Scientific Diagram

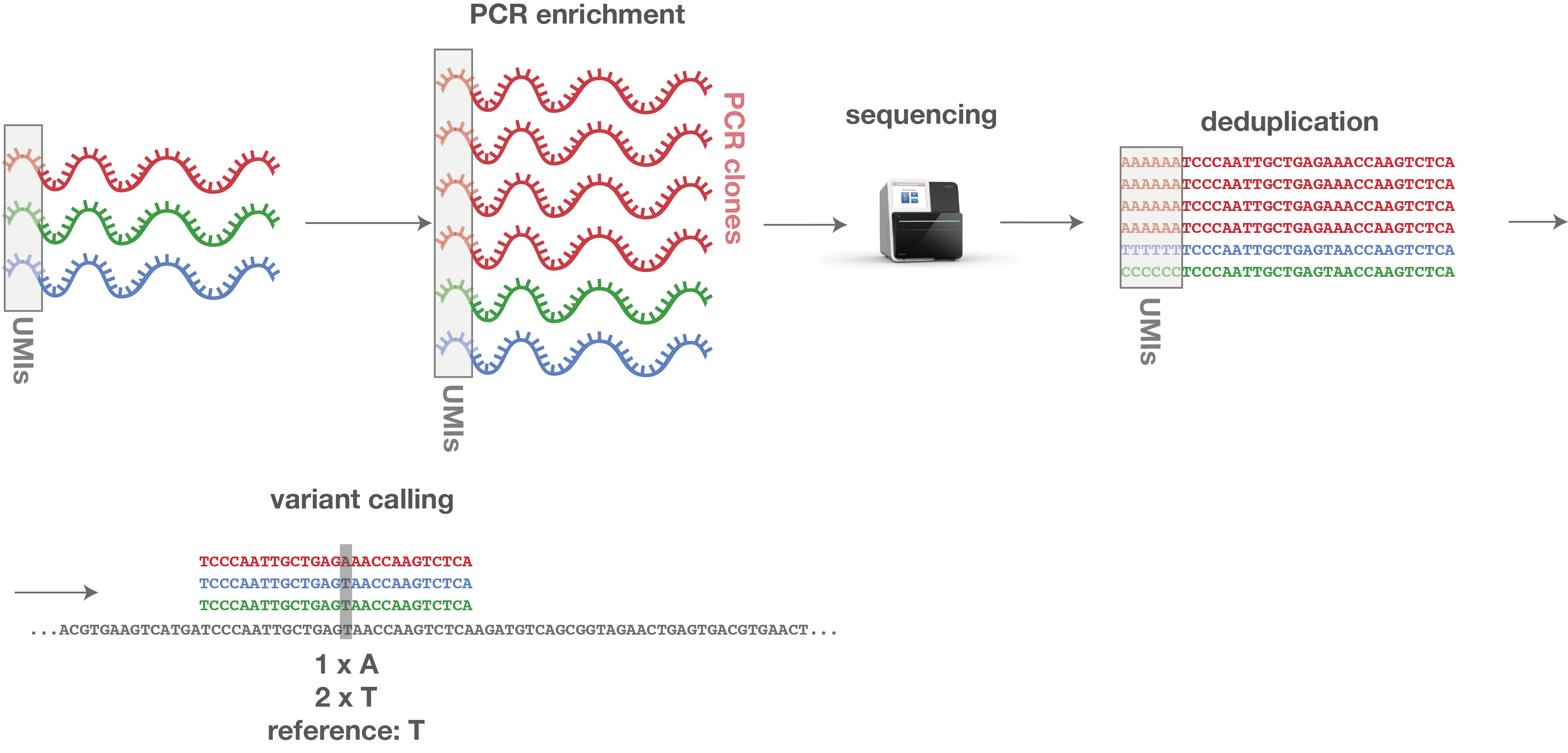
a This figure illustrates examples of various classes of UMI collisions... | Download Scientific Diagram

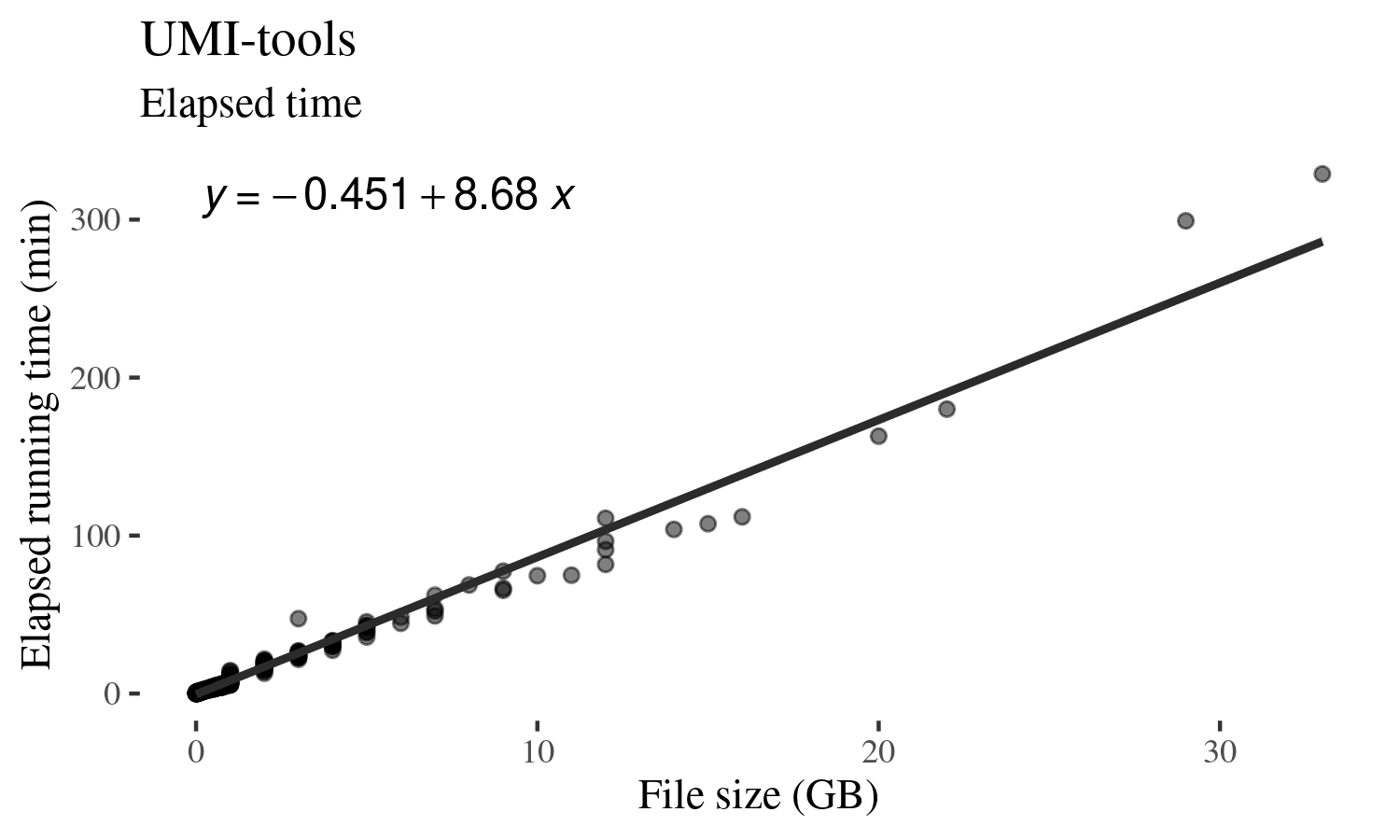
UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv